
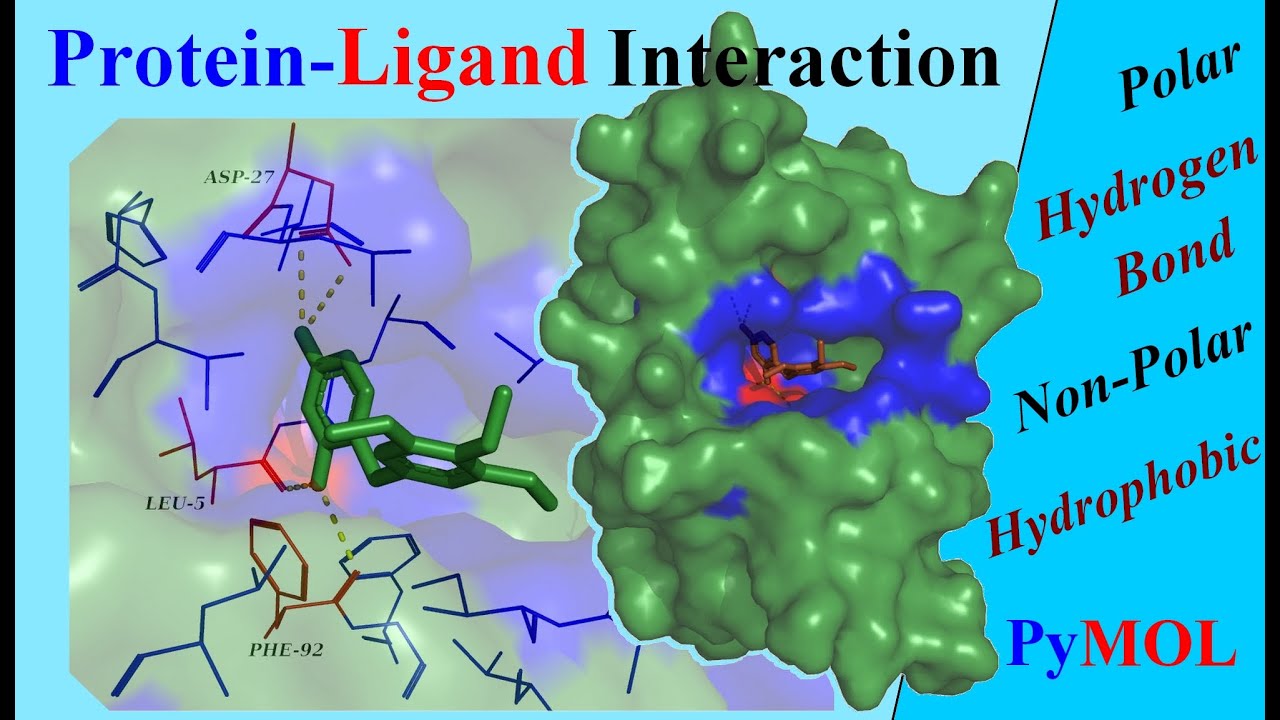
This protein is an enzyme called Triose Phosphate Isomerase and its structure is one of the superfolds we encountered in lecture 3. Now you know how to create a picture of a molecule indicating ligands bound and highlighting specific amino acids. The amino acids highlighted should be close to the ligand you have displayed. by displaying as ball and stick and giving particular colours. Select each of the amino acids you found when answering question 6 and highlight them e.g. We can apply the same process to highlight the individual amino acid residues which are involved in the active site. Once you have changed the display for the ligand, click on it once again to deactivate this selection. You could for example show it as spheres and give it a particular colour Now you can change the display of this selection (referred to by (sele) in the menu at the right hand side. This selects all the atoms in the ligand. It will also appear at the end of the sequence for the second polypeptide chain, but you need only change the display for the ligand bound to one protein chain.Ĭlick on the name for the ligand. It should be at the end of the sequence for the first polypeptide chain.
Pymol tutorial how to show ligand interactions code#
Scroll along the sequence display until you see the code for the ligand. Once this is done, it is possible to use this to select individual residues or ligands and modify the way they are displayed.
This will display the sequences of the polypeptide chains and the bound ligands above the display of the structure. Now select the Display menu at the top of the page and select the sequence option. At this stage you may just see the bound ligand as a tiny molecule - one bound to each polypeptide chain. Displaying the protein and its ligand within PyMOLĭisplay the molecule within PyMOL and use the Preset>Pretty mode to create a ribbon diagram.


 0 kommentar(er)
0 kommentar(er)
